Genomic patterns of malignant peripheral nerve sheath tumor (MPNST) evolution correlate with clinical outcome and are detectable in cell-free DNA
Isidro Cortes-Ciriano*1, Christopher D. Steele2, Katherine Piculell3, Alyaa Al-Ibraheemi4, Vanessa Eulo5, Marilyn M. Bui6, Aikaterini Chatzipli7, Brendan C. Dickson8, Dana C. Borcherding9, Andrew Feber10, Alon Galor11, Jesse Hart12, Kevin B. Jones13, Justin T. Jordan14, Raymond H. Kim15, Daniel Lindsay16, Colin Miller1, Yoshihiro Nishida17, Paula Z. Proszek10, Jonathan Serrano18, R. Taylor Sundby19, Jeffrey J. Szymanski20, Nicole J. Ullrich21, David Viskochil22, Xia Wang23, Matija Snuderl18, Peter J. Park7, Adrienne M. Flanagan*2,16, Angela C. Hirbe*9, Nischalan Pillay*2,16 and David T. Miller*3; for the Genomics of MPNST (GeM) Consortium Affiliates.
Author affiliations can be found in the full article online (linked below)
Abstract
Malignant peripheral nerve sheath tumor (MPNST), an aggressive soft-tissue sarcoma, occurs in people with neurofibromatosis type 1 (NF1) and sporadically. Whole-genome and multiregional exome sequencing, transcriptomic, and methylation profiling of 95 tumor samples revealed the order of genomic events in tumor evolution. Following biallelic inactivation of NF1, loss of CDKN2A or TP53 with or without inactivation of polycomb repressive complex 2 (PRC2) leads to extensive somatic copy-number aberrations (SCNA). Distinct pathways of tumor evolution are associated with inactivation of PRC2 genes and H3K27 trimethylation (H3K27me3) status. Tumors with H3K27me3 loss evolve through extensive chromosomal losses followed by whole-genome doubling and chromosome 8 amplification, and show lower levels of immune cell infiltration. Retention of H3K27me3 leads to extensive genomic instability, but an immune cell-rich phenotype. Specific SCNAs detected in both tumor samples and cell-free DNA (cfDNA) act as a surrogate for H3K27me3 loss and immune infiltration, and predict prognosis. SIGNIFICANCE: MPNST is the most common cause of death and morbidity for individuals with NF1, a relatively common tumor predisposition syndrome. Our results suggest that somatic copy-number and methylation profiling of tumor or cfDNA could serve as a biomarker for early diagnosis and to stratify patients into prognostic and treatment-related subgroups.
Data availability
Raw sequencing data has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under the dataset accession number EGAD00001008608. Ultra-low-pass whole-genome sequencing data of cell-free samples is available under controlled access data at https://doi.org/10.7303/syn23651229.
Correspondence and requests for materials should be addressed to David T. Miller (MD, PhD): david.miller2@childrens.harvard.edu
Code availability
The code used to process the raw data is available upon request.
The GeM Consortium represents the most thorough and intensive classification of MPNSTs. Our results suggest that accurate clinical pathology classification requires incorporation of genomic information.
In all cases, the early events in MPNST evolution involve biallelic inactivation of NF1, TP53 and CDKN2A, as well as mutations in PRC2 complex genes in a subset of cases.
We looked for possible targets of therapy like immune signature and gene fusions (has not been studied previously), and were not able to identify these potential targets
Our detailed analysis of the genomic architecture revealed distinct pathways of tumor evolution that can be identified through H3K27 methylation status.
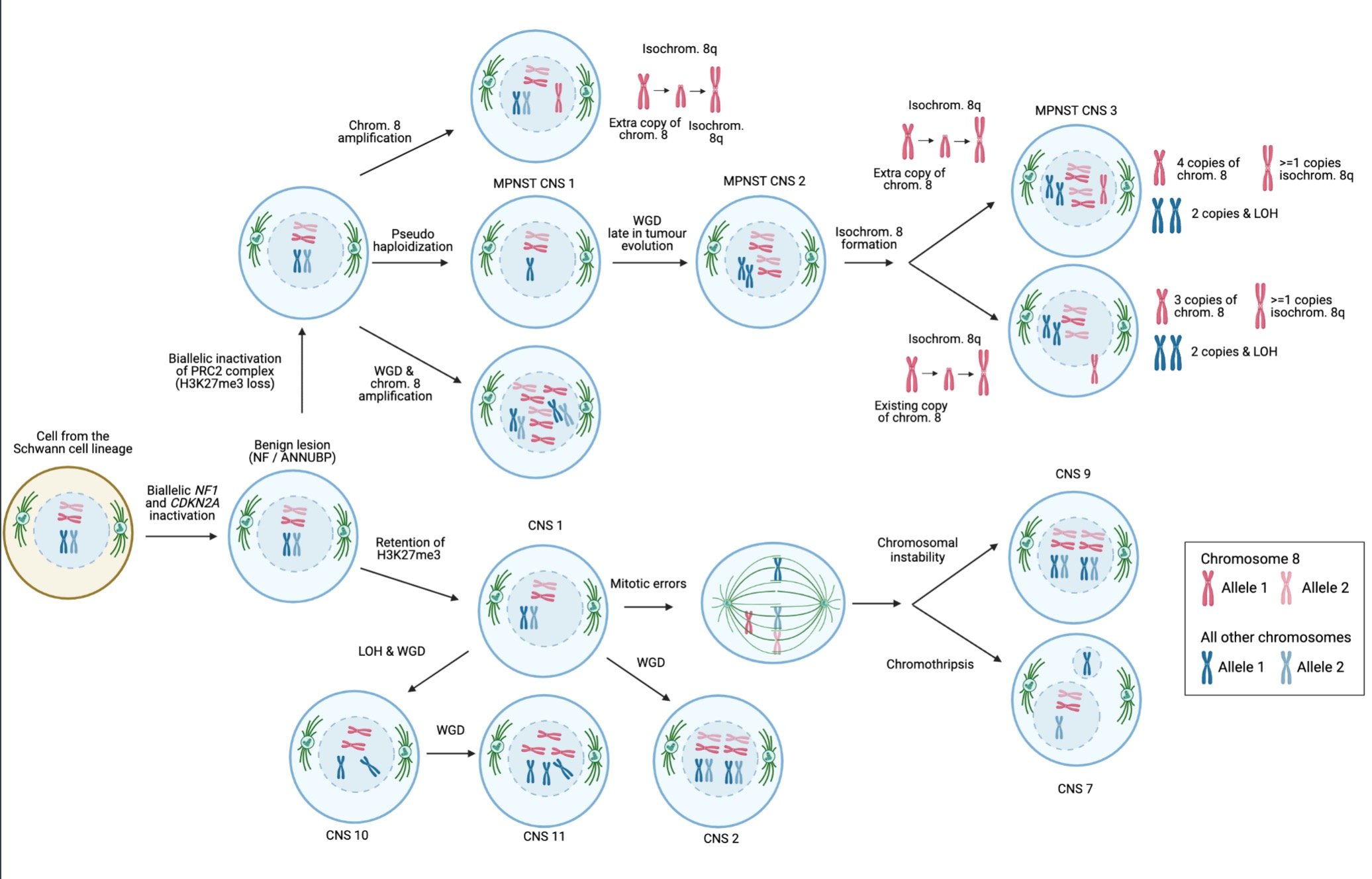
Pathways of MPNST evolution. Schematic representation of the order and timing of events involved in the evolution of MPNSTs. Copy number signatures predominant in specific karyotypic configurations are shown. CN, copy number signature; chrom., chromosome; isochrom., isochromosome; PRC2, polycomb repressive complex 2; WGD, whole genome doubling.
The NF Research Initiative at Boston Children's Hospital seeks to transform therapeutic development of NF-1 related Malignant Peripheral Nerve Sheath Tumors (MPNST) through international collaboration to improve the biological understanding of tumor progression in this disorder.
Partnership with the Park Lab at Harvard Medical School
The Genomics of MPNST (GeM) Consortium aims to advance preclinical research on targeted therapeutics for malignant peripheral nerve sheath tumors (MPNSTs), a rare cancer in patients with Neurofibromatosis (NF), through international collaboration and data sharing. To do so, we are building a controlled-access, cloud-based GeM Database, hosted by the Park Lab at Harvard Medical School. This data repository will host de-identified clinical and genetic data gathered from GeM collaborators, generated through comprehensive molecular analysis of tumor specimens, and produced through NFRI-funded research projects.
Dr. Park is an expert on genomic analysis related to cancer. Dr. Isidro Ciriano-Cortes, a former postdoc in the Park Lab, will be working with Dr. Park to conduct a genomic landscape analysis using whole genome sequencing and RNA sequencing data from MPNST specimen analyzed by the GeM Consortium.
The Park Lab has developed and hosted a database for the Pan-cancer Analysis of Whole Genomes (PCAWG) project that is similar in scope to the one envisioned for GeM. Dr. Park has agreed to construct the GeM Database to support data sharing and collaboration in the consortium and create an ongoing resource for future MPNST research.
An article titled “The Cancer Genome Atlas Pan-Cancer analysis project “ (Nature Genetics,2013), on which Dr. Park is a contributing author, describes this research initiative and the methods of comparison between tumor types profiled by The Cancer Genome Atlas. [Link to a PDF of the article here]